Options and Arguments of the SAXS Programs
*Version: 2005-10-12
*Introduction
*Prompts
*Arguments
*Options
*Examples
*Some Useful Commands
*Reference Systems
*Raster Orientation
*List of Options
*Example:
*Option Values
*Arithmetic Expressions as Values
*General Options Available in All SAXS Programs
*General Options for Output and Input Files
*Image Numbers and Memory Numbers
*File Access
*Loop Control
*Automatic Incrementation of File Numbers in a Loop
*Conversion between Multiple and Single Files
*Add Successive Images
*Transformation of Input and Output Images
*Image Dimension
*Reference System Parameters
*Replacement of Parameter Values with Header Values
*Error Propagation Options
*General
*Input/Output Data:
*Calculation
*Contents
*Storage
*Example
*SAXS Program Specific Options
*binary2saxs
*ccd2saxs
*gas2saxs
*gel2saxs
*saxs_add
*saxs_addcol
*saxs_addrow
*saxs_aff
*see also: saxs_rot
*saxs_ascii
*saxs_ave
*saxs_angle
*saxs_bsl
*saxs_cave
*saxs_clip
*saxs_col
*saxs_const
*saxs_csub
*saxs_div
*saxs_divcol
*saxs_divrow
*saxs_gnaw
*saxs_log
*saxs_mac
*saxs_mul
*saxs_mulcol
*saxs_mulrow
*saxs_normn
*saxs_patch
*saxs_pol
*saxs_power
*saxs_refract
*saxs_rot
*saxs_row
*saxs_scal
*saxs_stat
*saxs_sub
*saxs_subcol
*saxs_subrow
*saxs_tiff
*saxs_waxs
*rapid2saxs
*sphere2saxs
*mca2saxs
*Other Routines
*Programs
*col2array
*Macros
*saxsdisp.mac
*Error Propagation
*Mathematical Concept
*Expectation Value, Variance and Covariance
*Principle of Error Propagation
*Error Propagation During 2d-data Reduction
*Explicit Error Propagation Formulas
*Remapping of data arrays
*Scaling of two data arrays
*Summation of two data arrays
*Multiplication of two data arrays
*Division of two data arrays
*Index
* Options and Arguments of the SAXS ProgramsParameters can be passed in three ways to the saxs programs:
a) by prompts
b) by arguments
c) by options
The multiplication of the first three images of the input file "x.edf" with the factor 5.5 is used to demonstrate these possibilities. The output is written to "y.edf"
The program prompts for some important parameters, but not for all.
Usage: saxs_mac [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <ofac> <ocon> <shft1> <shft2>
saxs_mac saxs_mac 3.12 24-May-1995, Peter Boesecke
Input sequence 1 [input.edf] : x.edf <return>
Output sequence [output.edf] : y.edf <return>
First image of input sequence 1 [1] : <return>
Last image of input sequence 1 [1] : 3 <return>
Increment of input sequence 1 [1] : <return>
Output dummy [0.000000e+00] : -1 <return>
Output dimension 1 [1024] : <return>
Output dimension 2 [1024] : <return>
<output image> = <input image> * Factor + Const
Multiplication factor [1.000000e+00] : 5.5 <return>
<output image> = <input image> * Factor + Const
Addition constant [0.000000e+00] : <return>
Input reference system is Image
output shift 1 [0.000000e+00] :
Input reference system is Image
output shift 2 [0.000000e+00] :
...
...
The same parameters can be passed as arguments in the same order as the program would prompt for them. "=" stands for an empty argument. The option -p is used to switch off the prompt mode. With +p the program will prompt and will show the arguments as defaults in parenthesis []. This can be used for checks.
Usually, the default of all arguments can be set with options. While arguments must be passed in a specific order options can be passed in any order before the first argument. Options are preceeded by "-" or "+" and can be repeated. The last occurence sets the value.
If image1.edf and image2.edf contain several images the first image in image2.edf is subtracted from all images in image1.edf and written to output.edf.
More complex operations between images are possible, e.g. subtraction of series of background images from series of images. In these cases special options must be used.
In the following examples all header values of the input images are passed to the output images (+pass). The user will not be prompted (-p) and existing output files will be overwritten (-omod n). The command will be automatically applied to a series of input files.
In the first example the intensity of the input image is normalized to the header value Intensity1 (-scal) and then divided by by a flat-field image (+flat -i3nam flat.msk). The intensity of each pixel is divided by its spherical angle (calculated from the header values Center_1, Center_2, Psize_1, Psize_2 and SampleDistance). The output image is binned by 4 in each direction (-obin 4 4) and finally multiplied by 1.8/2.75 (-ofac 1.8/2.75).
This operation is done for all input files stp3_0002.edf to stp3_0039.edf (-i1nam stp3_%%%.edf,2,39). The results are written to the files stp3_0002.nrm to stp3_0039.nrm (-onam stp3_%%%.nrm).
If the values of center, pixel size and sample distance are not correct in the input images they can be set with the options -i1cen <Center_1[pc]> <Center_2[pc]> -i1pix <Psize_1[m]> <Psize_2[m]> -i1wvl <WaveLength[m]> (pc = pixel coordinate). Attention, the length unit is always meter.
The following command performs an azimuthal regrouping of the input image. Axis 1 of the output image corresponds to the radius, axis 2 to the azimuthal angle. The default creates 360 sections of 1 degree that are written to a row of the output image. The center and pixel sizes must be correctly defined in the header, otherwise the options shown above (-i1cen and -i1pix) must be used to update them.
This operation is done for all input files stp3_0002.nrm to stp3_0039.nrm (-i1nam stp3_%%%.nrm,2,39). The results are written to the files stp3_0002.ang to stp3_0039.ang (-onam stp3_%%%.ang). In this case the input and output files are passed as arguments in the same way the program would prompt for them.
The following command averages over all rows of the input image, in this case the azimuthally regrouped scattering pattern where each line corresponds to a sector. Like in the previous cases the operation is applied to all images between stp3_0002.ang stp3_0039.ang and the file names are passed as arguments. Each output image contains only a single row, the azimuthal average.
The last command converts the azimuthal average to a text file with two columns (+swap) that are separated by a tabulator (ASCII TAB character). The first column contains the scattering vector q in 1/nm, the second column the intensities. The default for scattering vectors is s=2sin(Theta)/WaveLength (in 1/nm). The option -scf "2*pi" multiplies the scattering vector by 2pi: 2*pi*s = q (in 1/nm). The scattering vectors are calculated using the scattering angle and trigonometric functions (+waxs) and not an approximation for small angles. The table is preceded by a summary of header values, e.g. the data history. Column labels (-headl "lab1 lab2 ...") are written above the columns. The first column label is "#q*nm", the second column label is "I([WaveLength]m)", where [WaveLength] will be replaced by the header value of WaveLength. The hash (#) is only added to allow a plot with gnuplot.
The output file looks like
#{
#Image = 1
#Size = 756
...
#History-4 = saxs_row +pass -omod n -p stp3_%%%.ang,2,39 stp3_%%%.row
#
#
#
#}
#q*nm I(1.75114e-10m)
0.0136098 -10
0.0408293 -10
0.0680488 -10
0.0952682 -10
0.122488 -10
0.149707 6.08817
0.176926 7.82687
...
Currently, seven different reference systems are defined: array, image, center, region, real, normal, saxs. They are used to define the geometrical relation between two images. A reference system is chosen with the option -rsys <refsys> (see chapter Option Values);

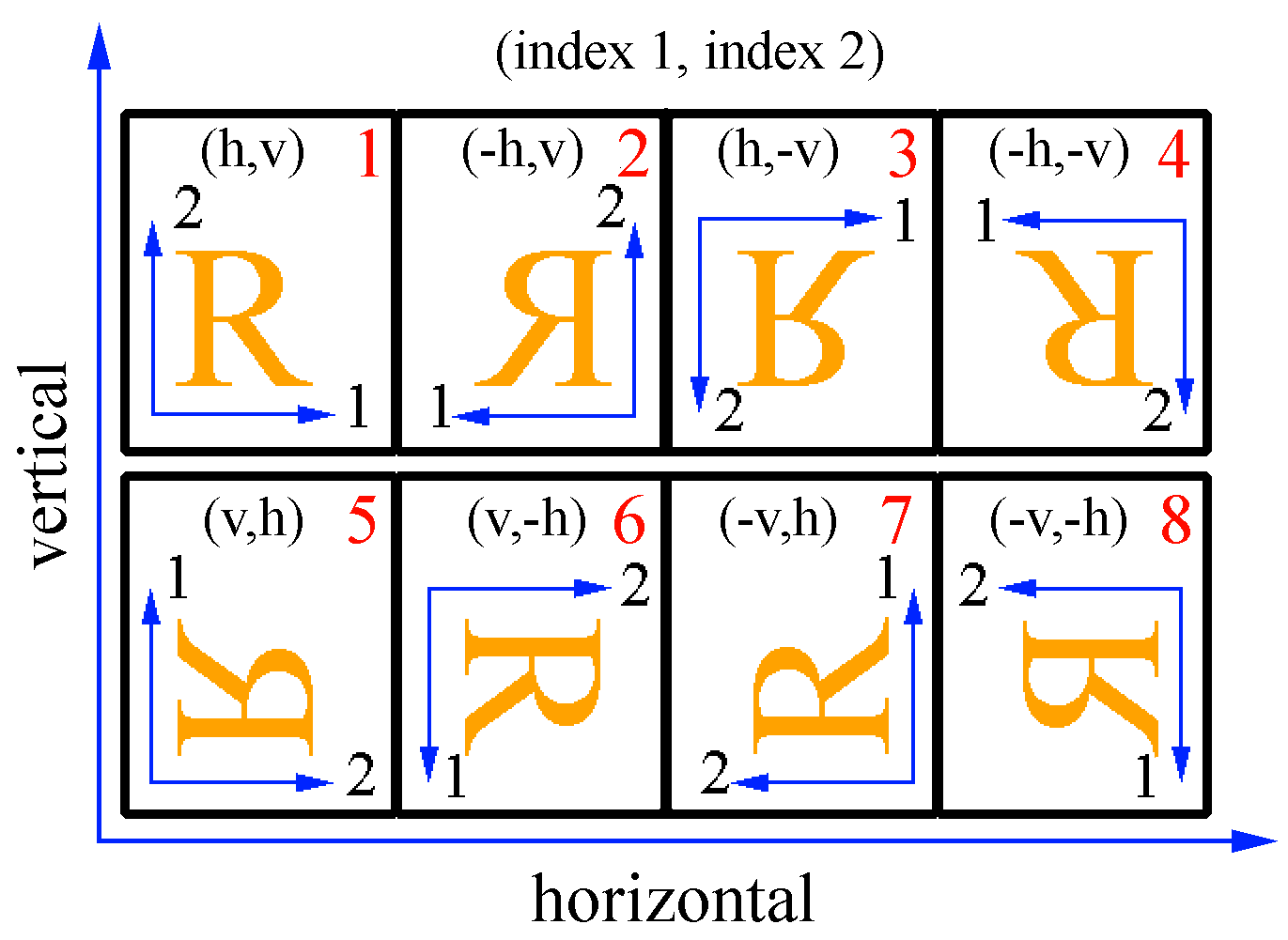
The orientation of the image describes the relation between data indices and directions in real space. In orientation 1 index 1 corresponds to the horizontal axis and index 2 to the vertical axis. Internally, all images are converted to orientation 1. The conversion includes a reorientation of the image and a swapping of some header values, e.g. for dimension, pixel size and offset. The orientation of the input and output image can be chosen with the options -i1ori O and oori O, where O is the orientation number shown in the upper right corner of Fig. 1.
Each option in the following list is followed by a parenthesis that shows the number of parameters and the parameter type:
option [<num of parameters>,<typ>]
The saxs programs are called in the following way:
saxs_xxx [option] {[option]} [argument] {[argument]}
Options are preceeded by '-' or '+'. Options are read from left to right
and are processed before the arguments are read. Options can be repeated.
The last option sets the value.
saxs_normn +flat -i3nam "flat_field" -trm 0.78 -ofac 1/0.0003 \
image.edf = = image.nrm
Each option can be followed by parameters. The number of parameters
and the type of the parameter are shown in brackets. The following parameter
types are used:
(Saxs, Waxs), e.g. -opro Waxs
(array, image, center, real, region, normal, saxs), e.g. -rsys saxs
Arithmetic Expressions as Values
Integer and float values can be expressed by C-type expressions. Values are converted to SI-units, e.g. keV is converted to Joule. Additional functions and constants are defined. To get a full list of all defined constants and units type "calc debug=2 a" on the command line. Units can be appended with an underscore to a number, e.g. 1_nm for 1e-9_m. The multiplication with a unit is only the multiplication with a conversion constant to a base unit, e.g. nm=1e-9, m=1.
Physical and mathematical constants are taken from:
[86] Lawrence Berkeley Laboratory
University of California
Berkeley, California 94720
X-Ray data booklet, second printing, with corrections, April 1986
Constants that are not contained in this edition are taken from
[91] Kuchling, Taschenbuch der Physik, Verlag Harri Deutsch 1991
There are currently some inconsistencies in some definitions, e.g. amu, ga and p.
gamma = 0.577215664901532861;
e = 2.71828182845905 /* Euler number */
k = 1.380662e-23; /* Boltzmann constant (J/K) */
me = 9.109534e-31; /* electron rest mass (kg) */
mp = 1.6726485e-27; /* proton rest mass (kg) */
NA = 6.022045e23; /* Avogadro number (1/mol) */
pi = 3.1415926535897932384626;
re = 2.8179380e-15; /* classical electron radius (m) */
c = 2.99792458e8; /* velocity of light (m/s) */
ec = 1.6021892e-19; /* electron charge (C) */
h = 6.626176e-34; /* Planck constant (J*s) */
b = 1e-28; /* barn (1/m^2) */
amu = 1.6605655e-27; /* atomic mass unit (kg) */
keV = ec*1e3; /* keV (J) */
eV = ec; /* eV (J) */
km = 1e3; /* km (m) */
m = 1.0; /* (m) */
cm = 0.01; /* cm (m) */
mm = 1e-3; /* mm (m) */
um = 1e-6; /* um (m) */
nm = 1e-9; /* nm (m) */
rad(x) = pi/180.0 * x; /* transformation deg to rad */
deg(x) = 180.0/pi * x; /* transformation rad to deg */
pi(x) = pi;
sin(x)
cos(x)
tan(x)
asin(x)
acos(x)
atan(x)
atan2(x,y)
sinh(x)
cosh(x)
tanh(x)
floor(x)
ceil(x)
abs(x)
exp(x)
log(x)
log10(x)
pow(x)
sqrt(x)
round(x) = floor( x + 0.5 ); /* rounding */
true = 1l;
false = 0l;
yes = 1l;
no = 0l;
(long int) x : transformation of x to long integer;
(round) x : rounding and transformation of x to long integer;
General Options Available in All SAXS Programs
An option can be followed one or more parameters. All options are described in the following way:
<option name> [<number_of_parameters>, <parameter type>]
switches a test modus on (+test) and off (-test)
displays long (+h) and short (-h) help
switches prompt mode on (+p) and off (-p)
sets the number of successive images that should be added (see Add Successive Images)
(default: 1)
do not use, undefined behaviour
defines the reference system (-rsys array | image | real | saxs). The default value is image.
defines the user reference system (-usys array | image | real | saxs). The default value is usys.
defines, whether the edf output file will be preceeded by a general
block (+gblk) or not (-gblk) (default: -gblk)
(see description of new edf format)
selects globally the input byte order of bsl files. All bsl files that are read by a single routine must have the same byte order, allowed values (not case-sensitive): low_byte_first, high_byte_first (default: internal byte order of the machine, e.g. i386 -> LOW_BYTE_FIRST, all others currently -> HIGH_BYTE_FIRST)
sets the maximum line width in the output file. Lines longer than mlw will be terminated with a backslash and continued in the next line, default: not set.
sets the minimum header size. Headers will be at least mhs bytes long.
General Options for Output and Input Files
The following options can be used for output and for input files. An option for the output file starts with o, an option for the <n>-th input file starts with i<n>, e.g. -odum -1 -i1dum -1 -i2dum -1 etc. The number of input and output files can be viewed with the extended help option +h (block should be read as file):
...
Number of image blocks : 3
Maximum number <n> of input blocks: 2
...
The file contains 1 output file (-onam <output file name>) and 2 input files (-i1nam <name of input file 1>, -i2nam <name of input file 2>).
Some conversion programs do not have standard input and output files, e.g. gas2saxs. In this case the number of image files ("blocks") is 0.
Image Numbers and Memory Numbers
Image numbers offer the possibility to store a sequence of (small) images in a single file. All images are numbered and can be stored in different memories. Image numbers and memory numbers are integer values. Positive memory values stand for image data, negative memory values for error data. Memory zero must not be used. The default input memory number is 1, the default output memory number is the input memory number.
How image numbers can be chosen is described in the section Loop Control. Memory numbers can be chosen with the options -i<n>mem and -omem.
memory number of images in output (input) file, e.g. -omem -1.
output (input) file opening mode, e.g. -i1mod o -omod n+ip: open an existing file for reading and create a new output file, protect the images of the output file
output (input) file name, e.g. -i1nam "input.edf" -onam "output.edf"
The file "/dev/null" is used as dummy. Used as input, a saxs_xxx programs runs as if it would read from a file. All parameters can be defined explicitly by options, e.g. saxs_mac -i1dim 512 512 -i1con 5 /dev/null output.edf will create an image with 512 512 pixels and write it to output.edf. Each pixel has the value 5. Output to "/dev/null" suppresses the output to a real file.
All saxs_xxx programs read images sequentially from several input sequences (i1, i2, ...) and write the resulting images sequentially to a single output sequence (o). Per default the output images are described in the same way as the images of the first input sequence, e.g. title, pixel size etc. are copied from the input images of the first sequence. Only a single output image sequence is created: (Output[i01], Output[i02], ...). The output image is a function of the input images: Output[i0] = Function(image1[i1], image2[i2], image3[i3], ..., imageN[iN]). Per default, the start number (ofst), the increment (oinc) and the last number (olst) of the output image sequence are the same as start number (i1fst), increment (i1inc) and last number (i1lst) of the first input sequence. Thus, normally the output image number is equal to the number of the image in the FIRST input file.
i1fst, i1inc, i1lst -> maxloop1 = max(1,(i1lst-i1fst+1)/i1inc)
...
iNfst, iNinc, iNlst -> maxloopN = max(1,(iNlst-iNfst+1)/iNinc)
ofst, oinc, olst -> maxloop0 = max(1,(i0lst-i0fst+1)/i0inc)
maxloop = Min(loop1, .. , loopN, loop0)
If the increment of the output image is 0, maxloop0 is set to infinity and it is only written once at the end of the other loops.
If the increment of an input sequence J is zero the loop is stopped after the first loop (maxloopJ is set to 1).
The loop is stopped when the first incrementation loop has finished. Also negative increments are possible.
ImageLoop(...) {
for (O=OFirst,I1=I1First,I2=I2First,...;
O<OLast,I1<I1Last,I2<I2Last,...;O+=OInc,I1+=I1Inc,I2+=I2Inc) {
Read(I1,I2,...);
Function((CmdBlk * ) pcb, (ImgHeadBlk *) ihb, (int *) pstatus );
Write(O); }
}
Missing images are skipped. All start images must exist.
first image number of output (input) file, e.g. -i1fst 10 -ofst 10
last image number of output (input) file, e.g. -i1lst 20 -olst 20
increment of image number of output (input) file, e.g. -i1inc 2 -oinc 2, if oinc (i<n>inc) is 0 the ouput image is only written once at the end of the loop (the input image is only read once)
Automatic Incrementation of File Numbers in a Loop
All saxs programs >= V4.0 allow incrementation of file numbers. The command
processes the files input001.edf, input002.edf, input003.edf, ... input010.edf and writes the results to the files output001.edf, output002.edf, output003.edf, ... output010.edf.
Numbers in a file name are marked with place holders (percent signs %%%%%). The range of file numbers must be indicated by parameters after the file name, e.g.
input%%%.edf,<first>,<last>,<increment>
The parameters are separated by commas. Numerical expressions are possible, e.g.
input%%%.edf,1,1+20,2
Omitted parameters are replaced by default values. If possible, they are copied from a preceding input parameter, e.g.
saxs_mac input%%%.edf,1,20,1 output%%%.edf,,,2
The output file numbers will start with 1 (copied), will be incremented by 2 and will stop after 20 (copied):
output001.edf, ouput003.edf, ... output019.edf
Conversion between Multiple and Single Files
Multiple files with a single data section (images) can be written into a single file with multiple data sections by writing the output file name without placeholders for numbers, e.g.
will write input001.edf .. input020.edf to output.edf (image 1 to image 20). If the input data files contain itself several images, only the first image in an input file or the one specified with -i1fst is written into the output file.
In the same way a single file can be split into multiple files by giving placeholders in the output filename, e.g.
will write all images of input.edf to numbered output files that contain only single images (default: image number 1).
Successive output images can be added before they are written. The option -add <n> will add n successive images (n>=1), e.g.
will add input001.edf to input003.edf and write the result to output001.edf,
will add input004.edf to input006.edf and write the result to output004.edf,
...
will add input016.edf .. input018.edf and write the result to output016.edf
Attention, the sum of input019.edf and input020.edf will be written to output020.edf. It will be the sum of only two images. Not all saxs-programs do really add the images, some may only output the last image. A test is necessary before using this option.
Transformation of Input and Output Images
After reading and before writing the pixel values of all images can be modified. The transformations are done in the following order after reading the input data and before writing to the output file. If the option is omitted nothing is done.
clip (-omin, -omax),
gnaw (-ogna),
rebin (-obin),
orientation (-oori)
transform (-ocon, -ofac))
Attention, each step can create new dummy pixels, e.g. -odum 10 -ofac 0 -ocon 10 would replace all output pixels with the value 10 which is the dummy value.
minimum value in output (input) image, e.g. -omin 0.01, lower values are replaced with dummies
maximum value in output (input) image, e.g. -omax 10000, larger values are replaced with dummies
constant is added to output (input) image value before saving, e.g. -ocon 1000 (-i1con 1000), adds 1000 to each non-dummy pixel of an image before saving (after reading). Multiplication and addition are done at the same time according to: value' = value * factor + constant).
factor is multiplied to output (input) image value before saving, e.g. -ofac 0.37 (-i1fac), multiplies each non-dummy pixel of an image with 0.37 before it is saved (after it was read). Multiplication and addition are done at the same time according to: value' = value * factor + constant).
rebinning factor of the output (input) images, e.g. -obin 2 2 rebins the output image by a factor 2. The image dimension is automatically adapted.
input: input image is transformed from orientation i<n>ori to
output: output image is transformed from orientation 1 to orientation oori
The value range of ori is from 1 to 8 (see Fig. 1)
rounding of the dummy area of the output (input) images, e.g. -ogna 2 4 enlarges each dummy pixel by 2 pixels horizontally and by 4 pixels vertically.
This feature is only available for program versions 2.50 and higher. It switches between averaging and addition of rebinned or resized pixels. The values are calculated from partial pixel areas. The averages are done by dividing the sum by the sum of the partials of all contributing pixels.
dimension of each output (input) image, e.g. -odim 720 1000 sets the output dimension horizontally to 720 and vertically to 1000. Unused pixels are filled with dummy. This option overwrites the header information of an input image.
In case of an input image this dimension is used instead of the value coming with the image.
dummy value of each output (input) image, e.g. -odum -1 sets the output dummy value to -1. The value 0 is never a dummy. A value is a dummy if (abs(value - dummy) <= ddummy) (with ddummy > 0.1). Dummy values are ignored in calculations and are replaced by the output dummy value. This option overwrites the header information of an input image.
In case of an input image this dummy value is used instead of the value coming with the image.
ddummy value, radius around a dummy value, in which the pixel values are ignored. This value is automatically set and must be always larger than 0.1. There should be no need to use this option. See odum. This option overwrites the header information of an input image.
In case of an input image this ddummy value is used instead of the value coming with the image.
output (input) image title, e.g. -otit "black sample"
In case of an input image this title string is used instead of the title coming with the image.
(reserved for time string)
The reference system for calculation is globally chosen with rsys. The reference systems are described in a separate document.
offset of the output (input) image (in pixel)
This option overwrites the header information of an input image.
In case of an input image this offset is used instead of the offset coming with the image.
center of the output (input) image (in pixel). This option overwrites the header information of an input image.
In case of an input image this center is used instead of the center coming with the image.
pixel size of the output (input) image (in meters). This option overwrites the header information of an input image.
In case of an input image this pixel size is used instead of the pixel size coming with the image.
sample distance in the output (input) image (in meters). This option overwrites the header information of an input image.
In case of an input image this distance is used instead of the distance coming with the image.
wavelength in the output (input) image (in meters). This option overwrites the header information of an input image.
In case of an input image this wavelength is used instead of the wavelength coming with the image.
detector rotation angles in labs space around the sample with constant sample distance, first rotation around axis1 in lab space, then around axis 2 in lab space and finally around axis 3 in lab space (unit: radian). This option overwrites the header information of an input image.
In case of an input image these rotations are used instead of the rotations coming with the image.
projection type of the output (input) image, values are "Saxs" or"Waxs".
In case of an input image this value is used instead of the projection value coming with the image.
Replacement of Parameter Values with Header Values
Replacement of Parameter Values with Header Values (2003-05-25)
The parameters of some options can contain keys between square brackets. The keys and the square brackets are replaced by the header values of the keys. If a key cannot be replaced the parameter string is not changed and will probably cause an error when the parameter is a number. Only input header values can be used. The keys are searched in the corresponding input file. If a program has several input files the input file number (block number) can be explicitly written after the key separated by a comma. For example, to divide all values of input file 2 by the header value of Psize_1 in input file 2 one can write: -i2fac "1/[PSize_1]". Header values after output file options, e.g. -ofac "1/[PSize_1]", are replaced by header values of input file 1. A comma separated number after a key specifies the input file number in which the key should be searched, e.g. -i2fac "1/[PSize_1,1]" will search PSize_1 in input file 1.
Parameters that allow replacement by header values are marked in the help list (+h) by D and F. Usually all string parameters (s) allow replacement of header values. Exceptions are filenames. The following list shows parameters in which header values are replaced:
-shft, -i1/-omin, -i1/-omax, -i1/-ocon, -i1/-ofac, -i1/-obin,
-i1/-ogna, -i1/-otit, -i1/-otime, -i1/-odim, -i1/-odum, -i1/-oddum,
-i1/-ooff, -i1/-obis, -i1/-opix, --i1/-ocen, i1/-odis, -i1/-owvl, -i1/-orot
+var/-var : create/do not create variance arrays
(default: create only, if i1 image comes with variance array)
If one of these options is used error propagation is switched on. The option +val is not needed.
The variances are propagated using the laws of Error Propagation. Cross correlations between pixels are ignored.
The variance array contains the variance of each pixel of the image array. The variance array has the same dimensions and orientation as the image array. The pixel [i,j] of the variance array contains the variance of pixel [i,j] of the image array. The variance header contains only keywords that describe an array. (Dim_1, Dim_2, DataType, ByteOrder, DataRasterConfiguration). The variance array does not use any additional keyword. If there are any, theyare ignored. The variance values are calculated after the calculation of the image values. Negative values in the variance array describe undefined variance values. The default value of a variance array is 0 (=exact).
Input and output variance arrays are only created if the first input image
If the option +var is set (default: not set) default variance arrays with variance zero are created for all input arrays. If the option -var is given no variance arrays are read/created and no error propagation calculation is done.
The variance array are stored in a second edf data block with the data block ID
EDF_DataBlockID = <image>.Image.Error[.<mem>].
The suffix .<mem> is omitted for memory=1.
For convenience, the data block of the variance array is written after the data block of the image and the header of the variance array of memory 1 contains additionally the keyword Image.Error = <image>, where <image> is the image number of the EDF_DataBlockID. The image header contains in this case the keyword Image = <image>.
To define initial variances for the input array sphere.edf the following option must be given:
It assumes that each pixel of sphere.edf contains the number of detected photons and that the errors follow Poisson statistics (var=s2=N). The option -i1err _i1val sets the variance to the pixel value (_i1val) of sphere.edf, _i1val is a variable. The following variables are defined:
_i1val: input pixel value
_i1err: input variance value
_oval: output pixel value
_oerr: output variance value
All variables start with an underscore.
Additional errors, e.g. var=1.5, can be added with.
To avoid negative variance values, the following command can be given:
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Version
V4.02 2002-04-06
Arguments
binary2saxs [options] <fnam> <onam> <i1dim1> <i1dim2> <fskp> <ftyp> <fend> <fdvo> <fras> <odum>
Options
number of bytes to skip
binary file name
binary type : type of a single element
(old type styles are also accepted)
byte swapping on/off (+fend: swap bytes, -fend: keep byte order (default))
HighByteFirst (default: machine byte order) overrides option fend
<DataRasterConfiguration> (default: 1)
<DataValueOffset> (default: 0)
Number of image blocks : 0
Arguments
ccd2saxs [options] <i/p file> <dummy> <first> <last> <inc> <o/p file> <o/p first> <i/p hm>
Options
extension of the edf output file (default: ".edf")
Number of image blocks : 0
Arguments
gas2saxs [options] <i/p file> <dummy> <first> <last> <inc> <o/p file> <o/p first> <i/p hm>
Options
extension of the edf output file (default: ".edf")
Number of image blocks : 0
Arguments
gel2saxs [options] <i/p file> <dummy> <o/p file>
Addition of two image sequences
Arguments
saxs_add [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Average columns between col1 and col2 in image sequence 2 and add the results to each column of image sequence 1
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_addcol [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Options
first column
last column
Average rows between row1 and row2 in image sequence 2 and add the result to each row of image sequence 1
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_addrow [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Options
first row
last row
Affine transformation of an image sequence
Tranformation matrix T
![]()
![]()
with coordinates W (input image), W' (output image)
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_aff [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2>
<t00> <t01> <t10> <t11> <t20> <t21>
Options
transformation matrix element T00 (default: 1.0)
transformation matrix element T10 (default: 0.0)
transformation matrix element T20 (default: 0.0)
transformation matrix element T01 (default: 1.0)
transformation matrix element T11 (default: 0.0)
transformation matrix element T21 (default: 0.0)
Transformation of an edf-file into ASCII format
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_ascii [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2>
<header> <swap> <head_1> <head_2>
Purpose: Transformation into ASCII format
Options: +-head +-swap +-hd1 +-hd2 -hdmk <marker> -ext <extension> +-noi +-waxs
Options
+-hist write/do not write history into header (default: +hist)
+-noi suppress/do not suppress image number extensions in output files (default: -noi)
+-waxs calculate/do not calculate the exact scattering vector for large scattering angles. If the projection type is Saxs 2sin(Theta) is calculated from tan(2Theta) to calculated the scattering vector. If the projection type is Waxs this transformation is not done.
The option +waxs sets the reference system to -rsys saxs. (default: -waxs)
-scf <factor> : multiplication of coordinates with a scale factor, e.g. to tranform s in nm to q=2*pi*s in Angstrom (-scf "0.2*pi") (default: 1.0)
-pref 'prefix' : specify the prefix of the ouptput file
-ext 'extension' : extension of the output ascii file (default: ".txt")
+-head : enables/disables the header printing
+-swap : enables/disables swap of rows and lines
+-hd1 : enables/disables the i_1 coordinate printing
+-hd2 : enables/disables the i_2 coordinate printing
+-abs : print absolut values of coordinates
+-spc : spacer between data columns, default: tabulator
Averaging of two image sequences
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_ave [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Transformation of an image from cartesian coordinates to polar coordinates with pixel interpolation and azimuthal averaging.

The integration is done on rings with thickness dr around the center. The default thickness is the minimum of the x_1 and x_2 pixel sizes in the chosen reference system. In the default reference system (Image) the default thickness is 1. In the following explanation it is assumed that the pixels are quadratic. The default values ensure that each pixel of the source image contributes to the result. Therefore, the radial default value (dr) should not be changed. The azimuthal default (da) can be changed.
Each ring is divided in subsections so that the azimuthal length of each subsection is less than the thickness dr. The integration uses the trapezoidal rule. Integration points are calculated for the center of each subsection by interpolation of the four neighbouring pixels. The weight of each pixel is proportional to the common cross section of a pixel with a squared pixel with size 1x1 around the interpolated point. (dashed pixel in Fig. 2).
x_1 of the output file corresponds to the radius
x_2 of the output file corresponds to the angle
The angle coordinate (x_2) uses always the real reference system (Offset, PixSiz). Center, WaveLength and SampleDistance are not used for coordinate 2 of the output image.
Version
4.10 2004-12-03
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_angle [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <odum>
<i1cen1> <i1cen2> <r0> <dr> <dim1> <a0> <da> <dim2>
Options
-r0 <minimum radius>: default 0.0
-dr <radial interval>: default is a value that corresponds to 1 pixel
-a0 <azimuthal start angle [radian]>: default 0.0_deg
-da <azimuthal interval [radian]>: default 1_deg
+-s2 This option is used to multiply the input data with the square of the radius (in rsys units) before transformation. This allows the direct calculation of the scattering power re^2*Q/V from the data by summation over i_1. (default: no multiplication)
The options asym and csym are used together with the option s2. They allow proper azimuthal averaging of intensities when the scattering pattern shows fiber symmetry and the fiber axis is lying in the detector plane. The angle of the fiber axis with the w_1 axis is defined by the option asym.
-asym <rotation of the symmetry axis relative to the w_1 axis [radian]>: default: 0.0_deg
+-csym switches on/off the multiplication of the input pattern with Pi/2*sin(Alpha-ASym), where Alpha is the angle of the data point from the w_1 axis and ASym the angle of the symmetry axis from the w_1 axis (default: no multiplication).
Transformation of an edf image sequence into bsl format (daresbury otoko)
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_bsl [options]
<i1nam> <output header name> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2>
Options
-hnam <output header name>: XNNMMM.EDF (default: automatically created from input filename)
+bsl : write into a single bsl binary file (default: -bsl)
Averaging of a point mirrored sequence with itself.
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_cave [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <center1> <center2>
Options
-cen1 <WCenter_1> : center 1 (input: user system coordinate, coordinate calculated according to first image)
-cen2 <WCenter_2> : center 2 (input: user system coordinate,coordinate calculated according to first image)
Sets all values above maxclip to maxvalue and all values that are smaller than minclip to minvalue. Works like -min and -max on the read data (after transformation and binning), but the excluded pixels are set to minimum and maximum (not to dummy).
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_clip [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <micl> <macl>
Options
-macl : maximum
-micl : minimum
Projection of a vertical band in a sequence of images to a single column. The output column is swapped to a line. The swapped column is written to a line of the output image.
see also saxs_row
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_col [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <col1> <col2>
Version
V4.50 (2002-06-02)
Options
first column of the vertical band (input: user system coordinate, coordinate calculated according to first image)
last column of the vertical band (input: user system coordinate, coordinate calculated according to first image)
first row of the input image (input: user system coordinate, coordinate calculated according to first image)
last row of the input image (input: user system coordinate, coordinate calculated according to first image)
multiply output with pixel size (+vint) (default: -vint)
Use the option -/+i1ave to select between averaging and summation of pixels. The option +/- vsum is not available any more.
aliased to --> saxs_mac
Subtraction of a point mirrored sequence from itself.
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_csub [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <center1> <center2>
Options
-cen1 <WCenter_1> : center 1 (input: user system coordinate, coordinate calculated according to first image)
-cen2 <WCenter_2> : center 2 (input: user system coordinate, coordinate calculated according to first image))
Division of two image sequences. To calculate the inverse of each image in an image sequence with the name x.edf type: saxs_div -i1fac 0 -i1con 1 -i1nam x.edf -i2nam x.edf
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_div [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Average columns between col1 and col2 in image sequence 2 and divide each column of image sequence 1 by the results
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_divcol [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Options
first column
last column
Average rows between row1 and row2 in image sequence 2 and divide each row of image sequence 1 by the result
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_divrow [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Options
first row
last row
Sets pixels in an ellipse around a dummy also to dummy. Similar to the general option +ogna <gnc> <gnr>.
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_gnaw [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <gnc> <gnr>
Options
horizontal radius
vertical radius
Logarithm (base 10) of the image. To calculate the logarithm to another base multiply the output with -ofac "log(<base>)/log(10)"
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_log [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2>
Mulitplication with a factor and addition of a constant to each image in a sequence. This routine performs a simple read and write of an image sequence. All general options are available and can be used to modify the values, e.g. to multiply with a factor and to add a constant: +ofac <fac> +ocon <con>. To convert an edf-file or bsl file to a standard edf-file with 512 byte block size and float binaries type saxs_mac old.edf new.edf or saxs_mac A01000.BSL new.edf.
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_mac [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <ofac> <ocon> <shft1> <shft2>
Multiplication of two image sequences.
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_mul [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Average columns between col1 and col2 in image sequence 2 and multiply each column of image sequence 1 with the results
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_mulcol [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Options
first column
last column
Average rows between row1 and row2 in image sequence 2 and multiply each row of image sequence 1 with the result
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_mulrow [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Options
first row
last row
Normalization of an image sequence to absolute intensities (+ccd for ccd, -ccd for delay line detector, default: -ccd)
Number of image blocks : 4
Maximum number <n> of input blocks: 3
Version
2004-10-31
Arguments
saxs_normn [options]
<i/p file> <scaler file> [<flatfield file>] <o/p file>
<first image> <last image> <inc>
<first scaler header> <inc scaler header>
[<flatfield image>]
<o/p dummy> <o/p dim1> <o/p dim2>
<transmission>
<scaler I0 mon.> <scaler exposure time> <scaler anode counts>
<2nd scaler I0 mon.> <2nd scaler exposure time> <2nd scaler anode counts>
General Description
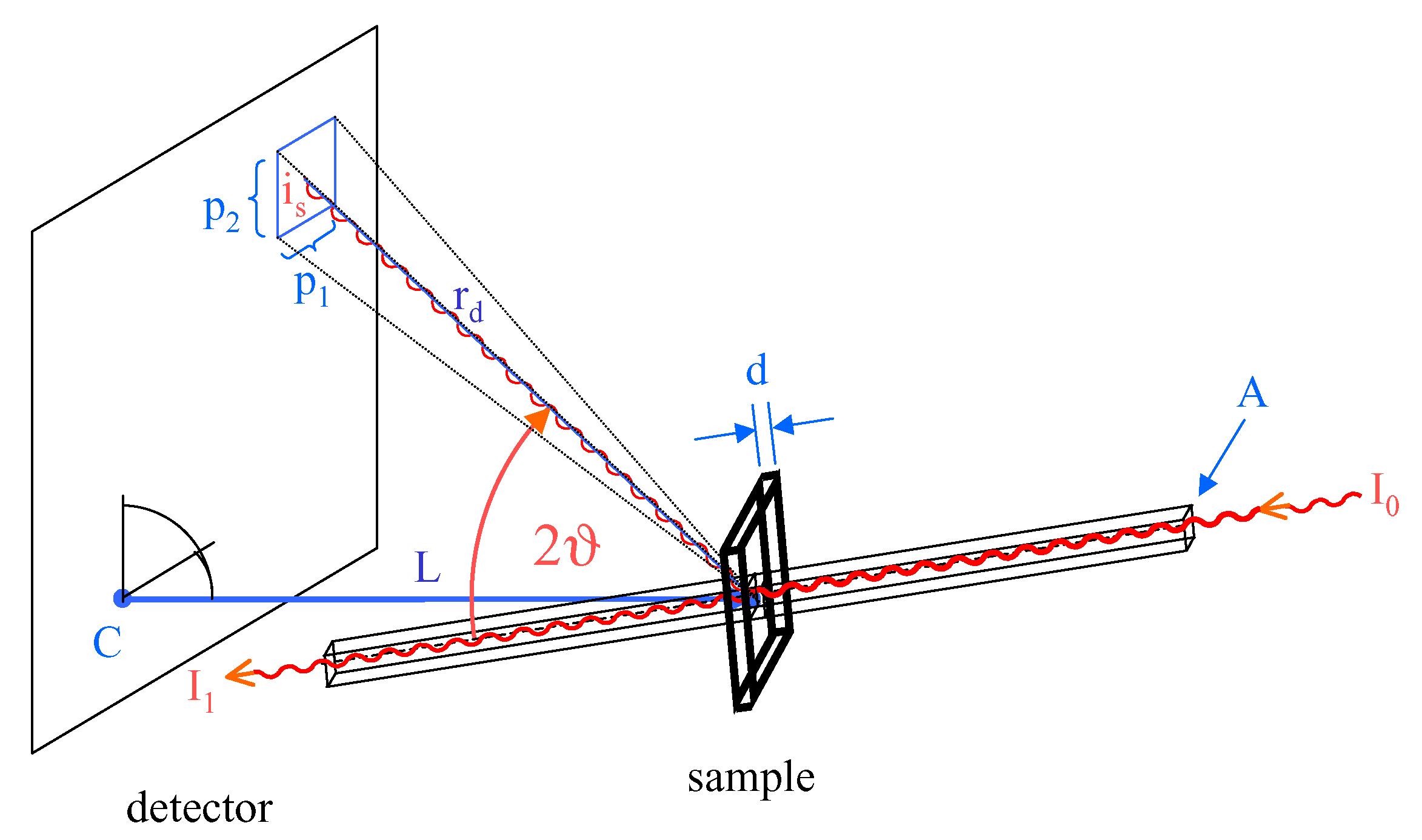
The program saxs_normn normalizes a two dimensional scattering pattern measured with a plane two-dimensional detector to absolute intensities in units of scattered photons per steradian and per incident photon. In other words, it is the scattering cross section ds/dW per sample cross section A. To correct for absorption the normalized pattern is divided by the sample transmission T. The last step is optional. The transmission corrected scattering cross section per sample cross section can be calculated in the following way:

I0 is the number of incident photons during the exposure, is is the number of detected photons in a single pixel with the area p1•p2, rd is the distance between sample and pixel and L the length of the detector normal to the sample (keyword SampleDistance). The division by the spherial angle dW is done by the last two factors in formula (1). The factor rd/L corrects the projection of the pixel area to the scattering sphere under large angles. To calculate the scattering cross section per scattering volume dS/dW of a non-absorbing sample the result must be divided by the effective sample thickness deff.
The division by the transmission T is effectively an absorption correction for a flat sample with thickness deff. In small angle X-ray scattering the transmission and the effective sample thickness are practically independent of the scattering angle. This is usually not the case in wide angle X-ray scattering. In this case the X-ray attenuation length and the sample shape influence the scattering pattern and formula (2) is no longer valid. In this case saxs_normn can still be used to calculate the number of scattered photons per steradian per incident photon, a property that can be measured without any knowledge about the scattering sample.
To take into account a non-homogeneous response over the detector area input pattern can be divided by a flat-field pattern F. A flat-field pattern is the detector response to a homogeneous illumination.
If extinction is small the transmission T can be approximated by I1/I0, where I0 is the number of photons in the incident primary beam during the exposure and I1 is number of photons in the transmitted primary beam.
Method
The program saxs_normn divides each pixel by the intensity I0 of the incident beam (option -monv <Intensity0>) and by the sample transmission. As default the transmission is equal to Intensity1/Intensity0, where Intensity1 is the number of transmitted photons (option -trmv <Intensity1>). Intensity0 and Intensity1 are read from the file header. The default corresponds effectively to a normalizion to Intensity1 because the Intensity0 cancels out. Alternatively, a fixed transmission value can be entered (option -trm <tranmission>). In this case, I1 is equal to T*I0. In this case, the header value Intensity1 is ignored. The pixel intensity is is divided by the spherical angle DW under which it observes the scattering.
To take into account the different sensitivities of the pixels the scattering pattern can be divided by a flat-field image F. In this case the option +flat must be set (default -flat) and the name of the flat-field image must be given after the option -i3nam <flat-field>.
The calculation of the transmission corrected scattering cross section per sample cross section is done in the following way:
is is the number of scattered photons at pixel coordinate (i_1, i_2), I(i_1, i_2) is the detector pixel intensity, F(i_1, i_2) is the flat-field value (+flat -i3nam <flat-field file> and f a normalization factor, that can be entered by the option -i1fac <f>)
T is the sample transmission, it can be modified with the option -trm <T>, Intensity0 can be modified with the option -monv <Intensity0> and Intensity1 can be modified with the option -trmv <Intensity1>
DW
is the spherical angle under which the detector pixel sees the scattering volume, PSize_1 and PSize_2 are the pixel sizes (option -i1pix <PSize_1> < PSize_2>), SampleDistance is the distance between detector and sample (option -i1dis <SampleDistance>). rd is the distance between the detector pixel and the scattering volume.(i_1,i_2) is the pixel coordinate, Offset_1 and Offset_2 are the coordinate offsets and (Center_1, Center_2) is the pixel coordinate of the point of normal incidence on the detector.
Call
The basic call to normalize a 2d CCD scattering pattern is:
a) without flat field
b) with flat field
The input file header must contain the following keywords. It they are missing the values must be given with the option shown in the second column:
|
Intensity0 |
-monv <Intensity0> |
|
Intensity1 |
-trmv <Intensity1> |
|
ExposureTime |
-timv <ExposureTime> |
|
Center_1, Center_2 |
-i1cen <Center_1> <Center_2> |
|
PSize_1, PSize_2 |
-i1pix <PSize_1> <PSize_2> |
|
SampleDistance |
-i1dis <SampleDistance> |
The parameter ExposureTime value is only required to display the detector summary. It is not used to calculate the output pattern. If not known, it can be set to 1. The displayed detector summary may be wrong.
If the input file does not contain the values Intensity0, Intensity1 and ExposureTime, they can be read from a second input file that contains the header values, e.g. the raw data file.
Data from a delay line detector
If the normalization is applied to data from a delay line detector the intensities can also be corrected for the dead time of the time to digital converter (TDC). In this case, the sum of the anode counts must be measured and given in the header after the keyword Anode (-anov <Anode>). The pixel intensities I(i_1,i_2) of the detector are scaled by the factor Anode/Sum(I(i_1,i_2)). In case of no losses the ratio should be 1. This correction does only work when the whole detector pattern is used, when the rejection is independent of the position on the detector and as long as the anode count rate is proportional to the global scattering intensity. Only global count rate losses are corrected.
|
Anode |
-anov <Anode> |
The basic program call is:
Multichannel scaler
It is possible to read the intensity information (Intensity0, Intensity1, Anode, ExposureTime) from a multi channel scaler (MCS). Normally, the program saxs_normn tries to find the multi channel scaler keywords HSI0, HSI1 and HSTime (and HSAnode in case of a delay line detector). If they are not found saxs_normn looks automatically for the keywords Intensity0, Intensity1, ExposureTime and Anode. The MCS counts of scaler nn is written after the keyword HS32Cnn, the zero value after the keyword HS32Znn, the calibration factor after the keyword HS32Fnn and its name after the keyword HS32Nnn. The scaler values are calculated in the following way:
and specifically:
To force saxs_normn not to read from scalers, the option -scal must be given.
Options
transmission (0..1)
eff. scaler depth in bits,
e.g. scaler depth=24 and 4 accumulations
=> -dpth "24+log(4)/log(2)" = 26
absolute counts of I0 scaler
zero value of I0 scaler
factor of I0 scaler (conversion of counts to photons)
number of incident photons
absolute counts of time scaler
factor of time scaler (for conversion of counts to seconds)
exposure time in seconds
absolute counts of anode scaler
factor of anode counter (for conversion of counts to anode counts, usually 1)
zero value of anode scaler
anode value in counts
absolute counts of I1 scaler
zero value of I1 scaler
factor of I1 scaler (conversion of counts to photons)
number of transmitted photons
absolute counts of second I0 scaler
factor of second I0 scaler (conversion of counts to photons)
absolute counts of second time scaler
factor of second time scaler (for conversion of counts to seconds)
absolute counts of second anode scaler
factor of second anode scaler (for conversion of counts to anode counts)
absolute counts of second I1 scaler
factor of second I1 scaler
sum of input image 1. This option can be used to set the sum of the input image externally.
The option -i1bin a b averages over a*b pixels. To calculate afterwards the correct rejection the binning correction factor must be set to -bcf "a*b". The binning correction factor cancels out and has no effect on the output image data, but only on the displayed acceptance and rejection values.
relative distance of sample from the home position of the detector displacement. If RelDis is set, the value of SampleDistance is ignored and replaced by the sum of DetectorPosition plus RelDis in input sequence 1 and 2 (i1 and i2) and in the output sequence (o). The value of DetectorPosition is read from input sequence 2.
number of I0 monitor scaler
number of exposure time scaler
number of anode counts scaler
number of I1 monitor scaler
number of second I0 monitor scaler
number of second exposure time scaler
number of second anode count scaler
number of second I1 monitor scaler
scaler length total number of available scalers (1..slen)
flatfield correction/no floatfield correction
Replacement of the first image by the second image (where it is defined). The second image is patched into the first image.
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_patch [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2>
Options
Division/multiplication by the polarization factor. All angles are in radian or must be followed by a unit, e.g 30_deg, 10_mrad.
The polarization factor P is splitted into an unpolarized P(1) and a polarized P(2) part:
The scattered beam kout is either calculated from an Ewald-sphere projection (ProjectionType = Waxs) or from a detector image (ProjectionType = Saxs). The calculation of kout is done with the same routines that are used in saxs_waxs. The rotation parameters etc are identical. Rotations are only used for detector images (ProjectionType = Saxs). The options -i1pro Saxs or -i1pro Waxs set the projection type manually.
The polarization state is described by the degree of polarization Pol and the Poincaré sphere: PChi (ellipticity ) and PPsi (polarization direction).
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Version
V4.52 2004-10-14
Arguments
Usage: saxs_pol [-mul]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <odum> <odim1> <odim2>
<pol[1.0]> <pchi[0.0]> <ppsi[0.0]>
Options
Calculate the n-th power of each image value
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_power [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <power>
Options
power exponent
Refraction correction of 2d-sub-surface scattering patterns. The calculation is valid for grazing incidence small angle scattering (GISAXS) and higher angle scattering. The incoming beam is refracted at the surface, scattered below the surface inside the sample and again refracted at the surface before it hits the detector. This routine transforms the observed (external) scattering pattern into the internal scattering pattern that would be observed inside the sample below the surface. The calculation is done using Snell's law. The surface normal N^ is defined by the angles chi and eta in the following way:
The primary beam is pointing against axis3 = axis1 X axis2. The surface normal N^ is initially pointing along axis1. It is first rotated ccw around axis3 (with chi). Then N^ is rotated around axis1 (with eta).
Version
V4.05 2004-10-14
Arguments
saxs_refract [options] <i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <delta[0.0_deg]> <the[0.0_deg]> <chi[90.0_deg]>
Options
1-delta : real part of the refractive index (0<=delta<1, default 0.0)
rotation of the sample around the primary beam in radian (corresponds to angle chi on id01) (radian or '_' and unit, 0_deg<=chi<=90_deg, default 90_deg). Chi is pi/2 (90_deg) when the sample surface is horizontal.
inclination of the chi-circle around axis 1 in radian (corresponds to angle eta on id01)
(0_deg<=eta<90_deg, default 0.0_deg)
Rotation of an image sequence by alfa in radian
![]()
![]()
with coordinates W (input image), W' (output image)
see also: saxs_aff
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Version
V4.04 2004-10-14
Arguments
saxs_rot [options] <i1nam> <onam> <i1fst> <i1lst> <i1inc> <odum> <odim1> <odim2> <alfa>
Options
ccw-rotation of the image-in radian. The rotation center is the origin of the chosen reference system.
Projection of a horizontal band in a sequence of images to a single row. The row is written to a line of the output image.
see also: saxs_col
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Version
V4.50 (2002-06-02)
Arguments
saxs_row [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <row1> <row2>
Options
first column of the input image (input: user system coordinate, coordinate calculated according to first image)
last column of the input image (input: user system coordinate, coordinate calculated according to first image)
first row of the horizontal band (input: user system coordinate, coordinate calculated according to first image)
last row of the horizontal band (input: user system coordinate, coordinate calculated according to first image)
multiply output with pixel size (+vint) (default: -vint)
Use the option -/+i1ave to select between averaging and summation of pixels. The option +/- vsum is not available any more.
Read the value of a keyword from the headers of each picture and write it to a text file.
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_scal [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <keyword>
Options
Keyword
Prefix of the output file
Extension of the output file
Name of the output file
Calculates statistics of the input image and write the result into a text file.
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_stat [options]
<i1nam> <i2nam> <o/p prefix> <i1fst> <i1lst> <i1inc>
<selection>
Options
selection
Prefix of the output file
Extension of the output file
Name of the output file
Print results to the standard output
Subtraction of two image sequences
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_sub [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Average columns between col1 and col2 in image sequence 2 and subtract the results from each column of image sequence 1
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_subcol [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Options
first column
last column
Average rows between row1 and row2 in image sequence 2 and subtract the result from each row of image sequence 1
Number of image blocks : 3
Maximum number <n> of input blocks: 2
Arguments
saxs_subrow [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc> <i2fst>
<odum> <odim1> <odim2> <i1con> <i1fac> <i2con> <i2fac>
<ocon> <ofac>
Options
first row
last row
Transformation of an edf image sequence to 1byte/2byte/4byte tiff files. If more than 1 image is read from the input file the output files are numbered. The input file is first converted to an intermediate output file and then converted to a tiff file. The output dummy is never scaled. The title is written as comment into the tiff file. The outptu byte order is given by the machine byte order.
Hint: To convert a 16 bit unsigned integer edf-raw data file to a tiff files with 16 bps the options "-auto -bps 16" should be set. If the file contains dummy values the output dummy value should be set to a positive value inside the mapping range. If it lies outside the output range it is forced to the closest number inside the output range.
Number of image blocks : 2
Maximum number <n> of input blocks: 1
Arguments
saxs_tiff [options]
<i1nam> <tiff file name -tnam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <8|16|32-bps> <1-8-rori>
<auto> [<min> <max>]
Options
bits per sample in output tiff image
<8|16|32> : 8, 16 and 32 bits in tiff image (default 16)
orientation of the output tiff image
1: (x,y)
2: (-x,y) x-axis inverted
3: (x,-y) y-axis inverted (default)
4: (-x,-y) both axes inverted
5: (y,x) axes swapped
6: (y,-x) axes swapped and x-axis inverted
7: (-y,x) axes swapped and y-axis inverted
8: (-y,-x) axes swapped and both axes inverted
-auto: As usual, the output dummy is set to -odum or its default. If not explicitely set by -min or -max, minimum and maximum of the input image are set 0 and pow(2,bps)-1, all values in the range [minimum, maximum] are mapped to [0,(pow(2,bps)-1)]
+auto: The output dummy is set to upper range value (pow(2,bps)-1). If not explicitely set by -min or -max, minimum and maximum of the input image are calculated, all values in the range [minimum, maximum] are mapped to [0,(pow(2,bps)-2)] (default +auto)
-auto should be used for format transformations, +auto for image display, e.g. with xv.
minimum of mapping range, values smaller or equal to min will be set to 0, all values in the range [minimum, maximum] are mapped to [0,(pow(2,bps)-1)]
maximum of mapping range, value larger or equal to max will be set to pow(2,bps), all values in the range [minimum, maximum] are mapped to [0,(pow(2,bps)-1)]
Example
saxs_tiff saxs_tiff 3.0 1999-11-12, Peter Boesecke
Input sequence 1 [input.edf] : flat.new230696
Output tiff file [flat.tif] :
First image of input sequence 1 [1] :
Last image of input sequence 1 [1] :
Increment of input sequence 1 [1] :
Output dummy [-1.000000e+00] :
Output dimension 1 [512] :
Output dimension 2 [512] :
Bits per sample (8, 16, 32) [16] :
Orientation of output image (1-8) [3] :
Range output image with minimum and maximum? [FALSE] :
Minimum value [0.000000e+00] :
Maximum value [6.553500e+04] :
input file : flat.new230696
output prefix : flat.tif
first image : 1
last image : 1
increment : 1
output dummy : -1.000000
output dimension 1 : 512
output dimension 2 : 512
bits per sample : 16
autorange : 0
minimum value : 0.000000
maximum value : 65535.000000
Reading image: flat.new230696, Image : 1 done
Calculating ihb[0, 1] = Function(ihb[ 1, 1] )
Input file : flat.new230696
Input image number : 1
Output tiff file : flat.tif
End of /home/boesecke/programs/saxs/bin/saxs_tiff
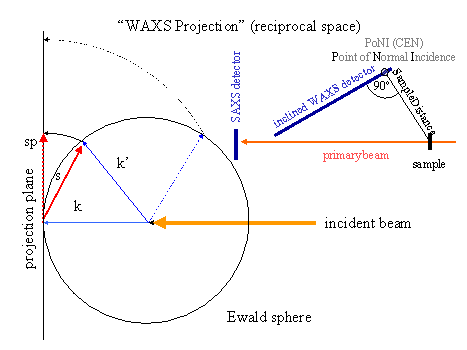
Projection of the Ewald sphere measured with a flat area detector (projection type Saxs) to a plane perpendicular to the incident beam (projection type Waxs) and vice versa. The detector can be inclined. Additionally, xy-displacement files can be written that can be used by the spatial distortion program spd. The projection will be called WAXS-transformation (see Fig. 4).
The length of the radial coordinate in the projected pattern corresponds to the length of the scattering vector s. The azimuthal angle corresponds to the azimuth of s. The projection is strictly valid for isotropic scattering patterns. In all other cases it is only a useful projection.
The scattering vectors are calculated using saxs-coordinates (default: -rsys saxs ). The keywords PSize_N, Center_N, SampleDistance and WaveLength are required. The detector rotation can be defined with the option -i1rot <i1rot1> <i1rot2> <i1rot3>.
Version
V4.12 2004-10-14
Arguments
saxs_waxs [options] <i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <i1rot1> <i1rot2> <i1rot3>
Options
An xy-displacement image contains for each input pixel the difference between the pixel position in the output array and the pixel position in the input array. The x-displacement file contains the displacements in x_1 direction, the y-displacement file contains the displacement in the x_2 direction. The xy-displacement images are defined for the input image. The displacement images have the same dimensions as the input image and they are described by the same geometrical header parameters.
The geometrical header parameters of the output image are written to the following keywords:
DisplacedBsize_1, DisplacedBsize_2
DisplacedCenter_1, DisplacedCenter_2,
DisplacedOffset_1, DisplacedOffset_2,
DisplacedProjectionType,
DisplacedPsize_1, DisplacedPsize_2
DisplacedSampleDistance, DisplacedWaveLength
They can be used by the spatial distortion program to write the header of the output image.
Example
Projection of a SAXS pattern to the Ewald-sphere
Conversion of rapid bsl files to edf files (frames, scalers, timing). The program uses the standard edfio routines to read rapid files.
It copies rapid data to edf data files including scaler data:
The general option -add <number of frames> allows summation over multiple frames. The scaler data are also summed. The timing data is also modified, the deltatime of the first added frame is used, the new exposure time is the sum of the exposure times of all added frames. rapid2saxs can also be used to average a sequence of edf-detector file and its header data (HS32Cnn, Intensity0 and Intensity1), e.g. a ccd file+ .
Example:
BSL data consist of a header file and a number of memory files. Each memory file contains three-dimensional binary data (rows, colums, frames).
The file name definition is very strict. Header and memory files must comply with the following naming convention (C: ASCII character, M, N: number between 0 and 9).
A rapid data set can consist of up to 4 files:
The header file defines the link between a file and a memory number. In the above example the header file looks like this:
SRS Data recorded on Sun Mar 23 15:43:27 2003
256 frames moving x-rays
512 512 256 0 0 0 0 0 0 1
A01001.323
256 1 1 0 0 0 0 0 0 1
A01002.323
256 4 1 0 0 0 0 0 0 0
A01004.323
The two first lines (maximum 80 characters) contain a description (free format).
The third and forth lines contain the format (rows, columns, frames) and the filename of memory 1, here A01001.323. The file contains 256 frames of 512 x 512 pixels. Each number consists of exactly 8 characters.
Each following successive line pair defines in the same way an additional memory. The last number is 1 except for the last memory definition.
The scaler memory (memory 2) contains 1 frame, 256 rows, each containing a single scalar value.
The timing memory (memory 3) contains 1 frame, 256 rows, each containing 4 scalars (dead time, live time, accumulated dead time, accumulated live time)
As a consequence of this the file numbers MMM of the memory files do not need to be consecutive. In the above example memory 3 is using the file A01004.323.
To rename a bsl data set it is necessary to update the file names in the header file.
Version
V4.53 (2003-04-15)
Arguments
rapid2saxs [options] <i1nam> <onam> <odum> <i2mem> <i3mem> <bibo>
Options
-i1nam <rapid header file> : frame data
-i1mem <memory of frame data, default 1>
-i1fst <first frame, default 1>
-i1inc <increment, default 1>
-i1lst <number of frames>
-i2nam <scaler data file, default i1nam> : scaler data
-i2mem <memory of scaler data, default 2> (-i2nam /dev/null" to ignore scaler memory)
-i2fst <first frame, default 1>
-i2inc <increment, default 0>
-i3nam <timing data file, default i1nam> : timing data (-i3nam /dev/null" to ignore timing memory)
-i3mem <memory of timing data, default 3>
-i3fst <first frame, default 1>
-i3inc <increment, default 0>
Other options:
-bibo <bsl input byte order, default high byte first>: 1 or 2
-add <number of images to add>
Generates the scattering cross section of diluted spheres
Version
2002-04-21 V4.04
Arguments
sphere2saxs [options] <onam> <odum> <odim 1> <odim 2> <opix 1> <opix 2>
<odis> <owvl> <rad> <dre> <vfr> <d><norm> <monv> <trm> <timv>
Options
Electron density diff. between particles and matrix [#/m^3] (default: 1e+28)
Particle radius [m] or distribution function (default: 1e-7)
Particle model:
The parameter "File" reads a table that must contain in colx the scattering vectors s in 1/nm and in coly the differential scattering cross sections. All lines starting with # are automatically skipped. With option -d <thickness> the tabulated values are multiplied by <thickness>.
colx: s=2sin(Theta)/lambda [1/nm]
coly: 1/V*(dSigma/dOmega) or 1/A*(dSigma/dOmega) for sample thickness d=1
Volume fraction (default: 0.01)
Sample thickness [m] (default: 0.0001)
Conversion of spec mca data to edf files (mca-frames, scalers). Only tested for spec files with one mca frame per scan
Version
V4.51 (2003-05-25)
Arguments
mca2saxs [options]
<i1nam> <onam> <i1fst> <i1lst> <i1inc>
<odum> <odim1> <odim2> <ofac> <ocon> <shft1> <shft2>
The scan number is chosen with i1fst.
Options
ignore the first skip scan numbers
(to access scans in the same file that have the same scan number)
A single column is extracted from a set of numbered ascii-input files and are rearranged to rows of a single ascii-output file. The columns must be separated by tabs ´\t´. The columns are numbered with 1, 2, 3 etc. from the start and -1, -2, -3 etc. from the end. The parameters must be entered in the described order. For omitted parameters default values are used.
Arguments
col2array [-h] [-b <mode[0]>] <output> <input> <first[1]> <last[1]> <inc[1]>
<col[1]> <skip_lines[0]> <skip_chars[0]> <separator['\t']>
Options
Example
The command
will read the last column of the files input_105.txt, input_102.txt, input_099.txt and input_096.txt and write it to output.txt. It will skip 2 lines and 0 characters at the start of each file.
Macros can only be used if the path to the saxs programs is defined in the shell initialization script (e.g. ".cshrc"). The initialization script must not change the directory.
A single image of an edf file is converted into 8 bps tiff format and displayed with xv. The output image is automatically scaled between 0 and 255. Dummy values are set to 255. The macro may be aliased to xdisp.
Arguments
saxs_disp <input> [<num>]
The error propagation is based on the following assumptions:
The variance values are stored in a separate variance array. Usually, the file size is doubled when error propagation is switched on.
Expectation Value, Variance and Covariance
The probability density f of a vector with n statistical variables xi can be written as
This can be written as the expectation value of an n-dimensional vector x:
The variance is the expectation value E of the squares of the differences between x and its mean value E[x]:
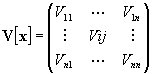
The matrix V[x] is called covariance matrix. It contains the variances Vii and the covariances Vij (i≠j).
Principle of Error Propagation
The input variables are described with x and the output variables with y.
The variance array of the output array V[y] is calculated from the variance array of the input arrays V[x] using the derivative matrix B. The input array x has n variables (pixels), the output array y has m variables (pixels).
The error propagation is calculated by transforming the input covariance matrix V[x] to the output covariance matrix V[y] according to the rule of error propagation given in eq. (24):
The matrix B contains the derivatives of each output variable (output pixel) yi with respect to each input variable xj (input pixel):
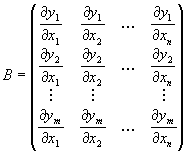
A straightforward application of this rule would result in an explosion of array sizes and calculation time. For practical purposes it must be simplified.
Error Propagation During 2d-data Reduction
Uncorrelated Input Data
In the case of uncorrelated input variables x the non-diagonal elements of the covariance matrix V[x] (eq. (21)) disappear and the diagonal elements V[y]ii of the output covariance matrix V[y] can be calculated with :
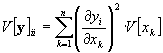
The non-diagonal elements of the output covariance matrix are
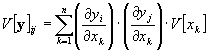
If the B matrix is not diagonal the output variables are correlated.
Pixel by Pixel Calculations
Identical number of input and output variables
If the number n and m of the input and output variables are equal the derivative matrix B can be diagonal. In this case the component V[y]ij of the output covariance matrix can be calculated directly from the component V[x]ij of the input covariance matrix:
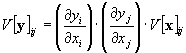
In this case the elements of the covariance matrix do not mix. The output variance array can be calculated independently for each variable, even if the input variables are correlated. This case corresponds to a pixel to pixel manipulation of a two dimensional data array, e.g. during scaling or normalization.
Smaller number of output variables than input variables
When L identically dimensioned input arrays with variables x1, x2, ... xL are combined pixel by pixel to an output array with the same number of variables the derivative matrix B can be splitted into L diagonal subarrays of the form:
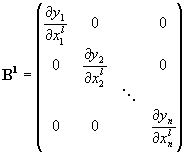
The matrix B can be written as.
The covariance matrix V can be written in the same way, if any correlation between variables of different arrays can be excluded.
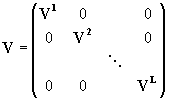
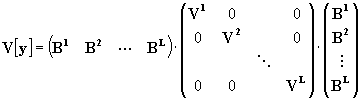
The component ij of the output covariance matrix is given by
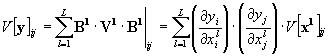
The summation is running over all input arrays l
Pixel by pixel combination of L identical input data arrays with uncorrelated variables
In the case that all input data variables are uncorrelated also the output variables are uncorrelated. The variance of each output data point can be calculated with eq. (33). In this case it is only necessary to store additionally to each output data value its variance, which is the diagonal value of the covariance matrix.
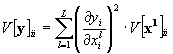
where V[y]ii = V[yi] = s2yi is the variance of data point i of the output array and V[xl]ii = V[xli] = s2xli the variance of the i-th data point of the l-th input data array.
Correlations between data points are not taken into account. This assumption is strictly valid if the calculations are done pixel by pixel. It is also valid if images are binned. It is generally not strictly valid if pixels are artificially divided into sub-pixels. This happens if an image is mapped to an image with different binning size. It leads to spatial smearing of the image.
Explicit Error Propagation Formulas
This operation writes the pixel intensities of region[i,j] of input array In1 into pixel Out[i,j] of the output array. A pixel has the area 1. The value of a partial pixel is calculated by multiplying the value of the full pixel with the partial pixel area.
A region is a rectangular area of an array. The sum of a region is the sum of all partial pixel values inside the region, the weight is the sum of all partial pixel areas inside the region. Dummy pixels are not take into account.
Sum(In1,region[i,j]): sum of all non-dummy partial pixel values in region[i,j] of array In1
Weight(In1, region[i,j]): sum of all non-dummy partial pixel areas in region[i,j] of array In1
a) Summation of data values
Out[i,j] = Sum(In1, region [i,j])
VOut[i,j] = Sum(VIn1, region [i,j])
b) Averaging of data values
Out[i,j] = Sum(In1, region [i,j])/Weight(In1, region [i,j])
VOut[i,j] = Sum(VIn1, region [i,j])/Weight(In1, region [i,j])^2
Out[i,j] = In1[i,j] * factor1 + constant1
VOut[i,j] = VIn1[i,j] * factor1^2
Out[i,j] = In1[i,j] + In2[i,j]
VOut[i,j] = VIn1[i,j] + VIn2[i,j]
Multiplication of two data arrays
Out[i,j] = In1[i,j] * In2[i,j]
VOut[i,j] = VIn1[i,j] * In2[i,j] ^2 + In1[i,j] ^2 * VIn2[i,j]
Out[i,j] = In1[i,j] / In2[i,j]
VOut[i,j] = (VIn1[i,j] + Out[i,j]^2 * VIn2[i,j]) / In2[i,j]^2